Epidemiology and Treatment
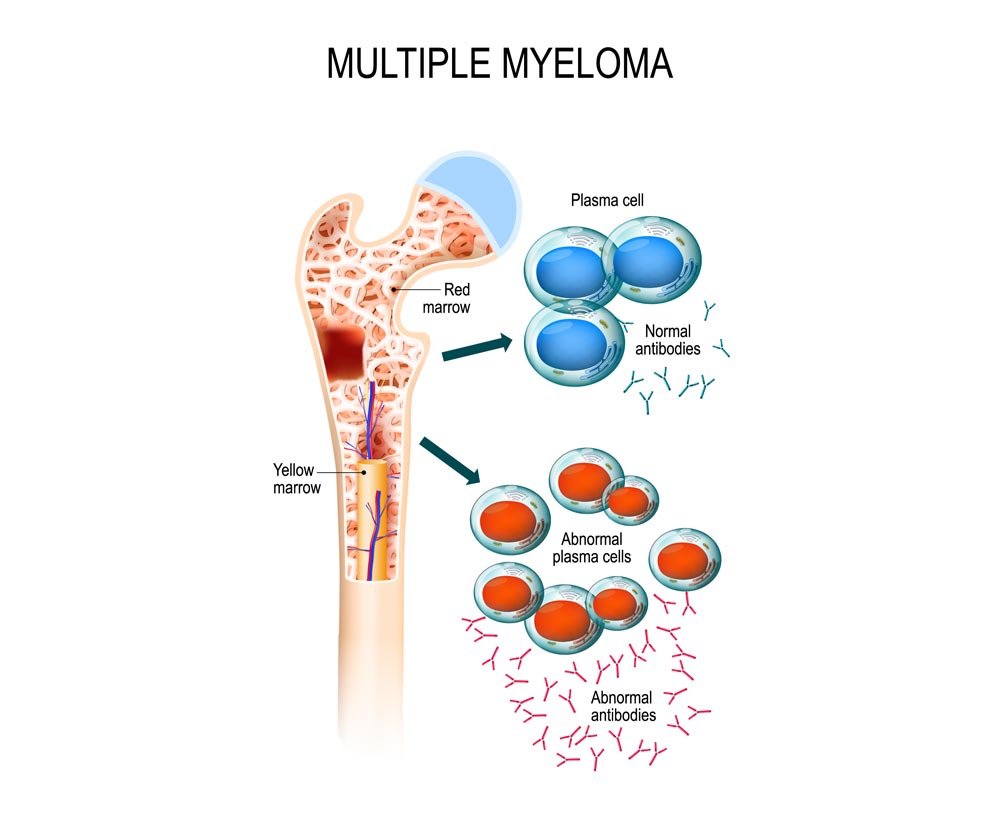
Multiple Myeloma (MM) is the second most common hematological malignancy in the world. It accounts for 1% of all cancers, 10% of all hematologic malignancies and 1 to 2% of all cancer-related deaths in Western countries. Each year, more than 25 000 new cases are diagnosed in Europe and the same number in the USA. MM is characterized by an abnormal clonal proliferation of malignant plasma cells within bone marrow and shows the significant complexity and genomic heterogeneity. The most typical clinical symptoms in myeloma are hypercalcemia, renal failure, anemia, and bone lesions.
Proteasome inhibitors, alkylating agents and IMID are effective therapies for myeloma and are become conventional treatments used alone or with other anticancer agents in MM at diagnosis. Studies showed that MM has the highest mean cost per case of all cancers involving the bone with around 100 000 €/year/patient.
Despite the introduction of new agents that have significantly improved clinical outcomes, most patients relapse and myeloma remains again incurable disease in the majority of cases. The wide inter-individual variation in response to treatment of patients with MM is the major limitation to achieve a consistent therapeutic effect.
Heterogeneity in Multiple Myeloma
Recent advances in next generation of sequencing and single cell genome profiling studies showed the extensive heterogeneity with clonal and subclonal levels. Therefore, novel therapeutic strategies to overcome chemotherapy resistance are needed in clinical settings. With the development of new biomarkers that can distinguish drug-sensitive and drug-resistant cancer cells, Diag2Tec answers to unresolved challenges and aims to develop personalized treatments in MM.
The study of multiple myeloma biology cells is not easy because plasma cells are in a bone marrow niche and interactions with the microenvironment are essential for their proliferation and survival. This is even more difficult for normal plasma cells because they are rare and difficult to harvest; they represent approximately 0.5% of mononuclear cells. It is from this perspective that our team has developed different unique models to study the biology of the multiple myeloma and offers you a large range of customized services.


